Experiment 3.1 Protease Structure and Function (updated: 02/09/99)
Background
In living systems, nearly all chemical transformations are catalyzed and regulated by enzymes. Enzymes allow chemical reactions to proceed (i) very quickly, at rates many orders of magnitudes greater than uncatalyzed rates, (ii) at mild temperatures in water, at neutral pH, (iii) with very high specificity and no side reactions, and (iv) with regulated rates, that are controlled by environment. Kinetic analysis of an enzymatic reaction is used to study substrate-enzyme affinity, the catalytic rate constant, and mechanism of catalysis and inhibition. Detailed kinetic information is necessary for understanding enzyme specificity.
Proteases are enzymes that hydrolyze amide bonds of proteins and peptides. Proteases play central roles digestion, blood coagulation, complement activation, fibrinolysis, apoptosis, mitotic signaling, and fertilization (1). There are four classes of proteases: (i) thiol proteases, which use cysteine as a nucleophile (papain coordinates), (ii) acid proteases, which use aspartic or glutamic acid as a nucleophile (hiv protease coordinates), (iii) metalloproteases (carboxypeptidase coordinates), and (iv) serine proteases, which use the hydroxyl oxygen of serine as a nucleophile (trypsin coordinates).
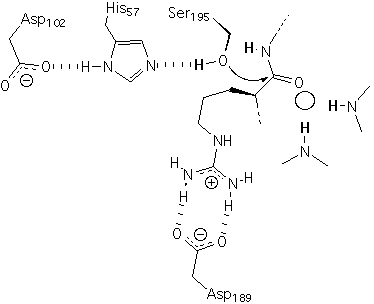
For serine proteases such as trypsin, which you use in this lab, the O gamma of serine 195 is the nucleophile that catalyzes amide hydrolysis (Figure 1). Serine 195, histidine 57 and aspartic acid 102 are linked by hydrogen bonds and act in concert during catalysis. These three residues are called the catalytic triad. This catalytic triad has such profound utility that it originated independently several times during evolution (i.e., by convergent evolution). Details of the mechanism can be found in Voet and Voet and several additional sources (2, 3).
Trypsin is produced in the pancreas and is secreted into the small intestine, where it hydrolyses amide bonds of proteins during digestion. In this lab you will be using bovine beta-trypsin. "beta" refers to a chromatographic property; the intact enzyme elutes as a second peak on an ion-exchange column. The first peak ("alpha") is a cleaved form of the enzyme. Cleavage occurs by autolysis - a process in which one trypsin molecule hydrolyzes an amide bond of another trypsin molecule. Commercially obtained trypsin is usually predominantly the "beta" (uncleaved) form with a minor "alpha"contaminent.

Figure 1: The catalytic triad, primary specificity pocket, oxyanion hole of trypsin. An arginine-containing substrate is bound in a Michaelis Complex
Proteases are specific. A protease will hydrolyze only particular amide bonds, depending on the identity of the amino acids before and after the scissile bond (the bond to be hydrolyzed). This specificity is defined by the following scheme (4).
+H3N-S3-S2-S1-cut-S1'-S2'-S3'-COO-
Figure 2: Scheme describing the protease specificity. Sn refers to residues on the amino terminal side of the scissile bond; Sn' refers to residues on the carboxy terminal side of the scissile bond.
Trypsin has the following specificity:
S3 = slight specificity for aromatic residues
S2 = slight specificity for proline.
S1 = strong specificity for basic residues.
S1' = no residue specificity.
S2' = no residue specificity.
S3' = no residue specificity.
The mechanism and class of a serine protease are essentially independent of the specificity. For example both a-chymotrypsin and trypsin are serine proteases, with nucleophilic serine residues and conserved catalytic triads. Yet the S1 specificity of a-chymotrypsin differs from that of trypsin; a-chymotrypsin has S1 specificity for bulky aromatic residues (phenylalanine) while trypsin has S1 specificity for basic residues (arginine and lysine).
The goal of this excercise is to use RASMOL to explore the active site and enzymology of a serine protease. Each student will study a different structure.
a) On the PDB homepage, under SEARCH on the right side of the page, go to "Search Fields: advanced search".
b) Under "Customize the search fields on this query form", click resolution. Do not make any other changes. Then click, "NEW FORM".
c) In the TEXT SEARCH field, enter "###### and $$$$$$", where ###### is the serine protease of your choice (thrombin, chymotrypsin, elastase, etc) and $$$$$$ is the organism of your choice [cow (bovine, Bos Taurus), human (Homo Sapien), Turkey (Meleagris Gallopavo), Pig (porcine, Sus Scrofa), Salmon (Salmo Salar), Bitter Gourd (Momordica Charantia), Rat (Rattus Norvegicus) etc.]. In the RESOLUTION field, enter LESS THAN OR EQUAL TO 1.8 Å. You might have to increase this number to 2.0, then 2.2, to get any hits). A smaller number in the resolution field (in Å) indicates greater accuracy and higher resolution. Click SEARCH. EXPLORE one of the protease entries that you pull up. Check that the molecule is a protease, and not simply a protease inhibitor, before proceeding too far. Save the pdb file to your disk. Open the file with a word processor. Email the first 10 lines of the PDB entry to the instructor/TA. In the subject line put "yourlastname, yourfirstname, CHEM 4581, protease pdb entry=xxx" Where xxx is the pdb entry number.
d) Open your protease file in Rasmol. Make several beautiful figures of the global structure of the protein and of the active site. Fortunately people who study serine proteases always use the same numbering scheme for the catalytic triad (Figure 1). The catalytic serine is always number 195, etc. Make a beautiful figure of the catalytic triad. Label the residues. Draw the hydrogen bonds. The structure probably contains an inhibitor bound in the catalytic site because a protease has to be inhibited during the crystallization or it will self-hydrolyze. Include the inhibitor in the figure.
e) Measure interaction distances between the residues of the catalytic triad and between the protein and the inhibitor, or any solvent molecules contained within the active site. Neglect distances greater than 3.5 Å because it is greater than the sum of the van der Waals radii of the atoms. Make a table with five columns: atom 1, residue 1, atom 2, residue 2, distance. Indicate which interactions are hydrogen bonds.
References (top=>)
1) Neurath, H. "Evolution of proteolytic enzymes." Science 224, 350-357 (1984).
2) Voet, D. Biochemistry, 2nd Ed., John Wiley and Sons (1995)
3) Creighton, T.E., Proteins, 2nd Ed., W. H. Freeman and Co. (1993)
4) Schechter, I., and Berger, A. "On the size of the active site in proteases. I. Papain." Biochem. Biophys. Res. Comm., 27, 157-162.